What Is BrAPI?
The Breeding API specifies a standard interface for plant phenotype/genotype databases to serve their data to crop breeding applications. It is a shared, open API, to be used by all data providers and data consumers who wish to participate. Initiated in June 2014, the BrAPI community has grown to over 200 software developers and agricultural scientists from 36 international institutions. A wide network of interconnected software has begun to emerge as more groups use BrAPI as the common language for communicating data.
The BrAPI specification is free and open source, and the BrAPI community is very welcoming and knowledgeable. All the specification documentation can be found on brapi.org and the BrAPI Getting Started Guide guide is a great place to start. To become part of this community, consider joining the mailing list, connecting on Slack, or following the project on Twitter.
Definitions
- BrAPI
The Breeding API (BrAPI) is a Standardized RESTful Web Service API Specification for communicating Plant Breeding Data. BrAPI allows for easy data sharing between databases and tools involved in plant breeding.
- API
An Application Programming Interface (API) is used to interact with and control an application programmatically. Just as a User Interface (UI) allows a human user to interact with an application, an API allows a separate piece of code to interact with an application. An API can be used to send input data, receive output data, configure settings, and trigger actions within an application.
- Web Service
Web Services are computational services accessed over a computer network. Typically, when people talk about web services, they are referring to a standardized web service communication protocol. The two web service protocols which are currently the most popular are SOAP and REST. The BrAPI project uses REST as the standard protocol for the specification.
- REST
Although the above paragraph indicates REpresentational State Transfer (REST) is a web service protocol, strictly speaking, it is an architecture or design framework. The REST architectural ideas are applied by using the HTTP protocol for information transport. REST uses the standard HTTP verbs (GET, POST, PUT, etc) to define different types of actions, and URLs to indicate where a request should be sent and what input data to use. When sending or receiving data, complex data structures are typically defined using JSON or XML.
- Standardized
BrAPI is an attempt to standardize the commands and data structures needed for typical plant breeding applications. The BrAPI Community is a key part of the project, bringing together people and technical resources from around the world to agree on a global standard for this kind of data. The standard is maintained and regularly updated, taking into account new use cases and data structures as the community grows.
- Specification
BrAPI is a technical Specification that software developers can easily turn into code and communicate data using the community standard models. There is a common misconception that BrAPI is a separate tool or an application, when in-fact each group must build their own BrAPI implementation. At its core, BrAPI is just a set of documentation that defines a standard for communicating data. However, there are many resources available within the BrAPI community that make it easy to build an implementation and make your system BrAPI compliant.
Core Documentation
The core BrAPI documentation can be found on the Specification page. From there you can find links to all the different versions of the standard. You can view the documentation on GitHub, Apiary, or SwaggerHub depending on your personal preference of presentation style. On GitHub, you will find the specification presented as README files, as well as the raw OpenAPI 3 formatted YAML files. Apiary is much more focused on text documentation and is often easier to read, but might be lacking in technical detail. SwaggerHub is more focused on technical detail, but human readable text definitions may be harder to find. SwaggerHub is also more interactive with a dummy server implementation to play with and a code generation tool in a variety of common programming languages.
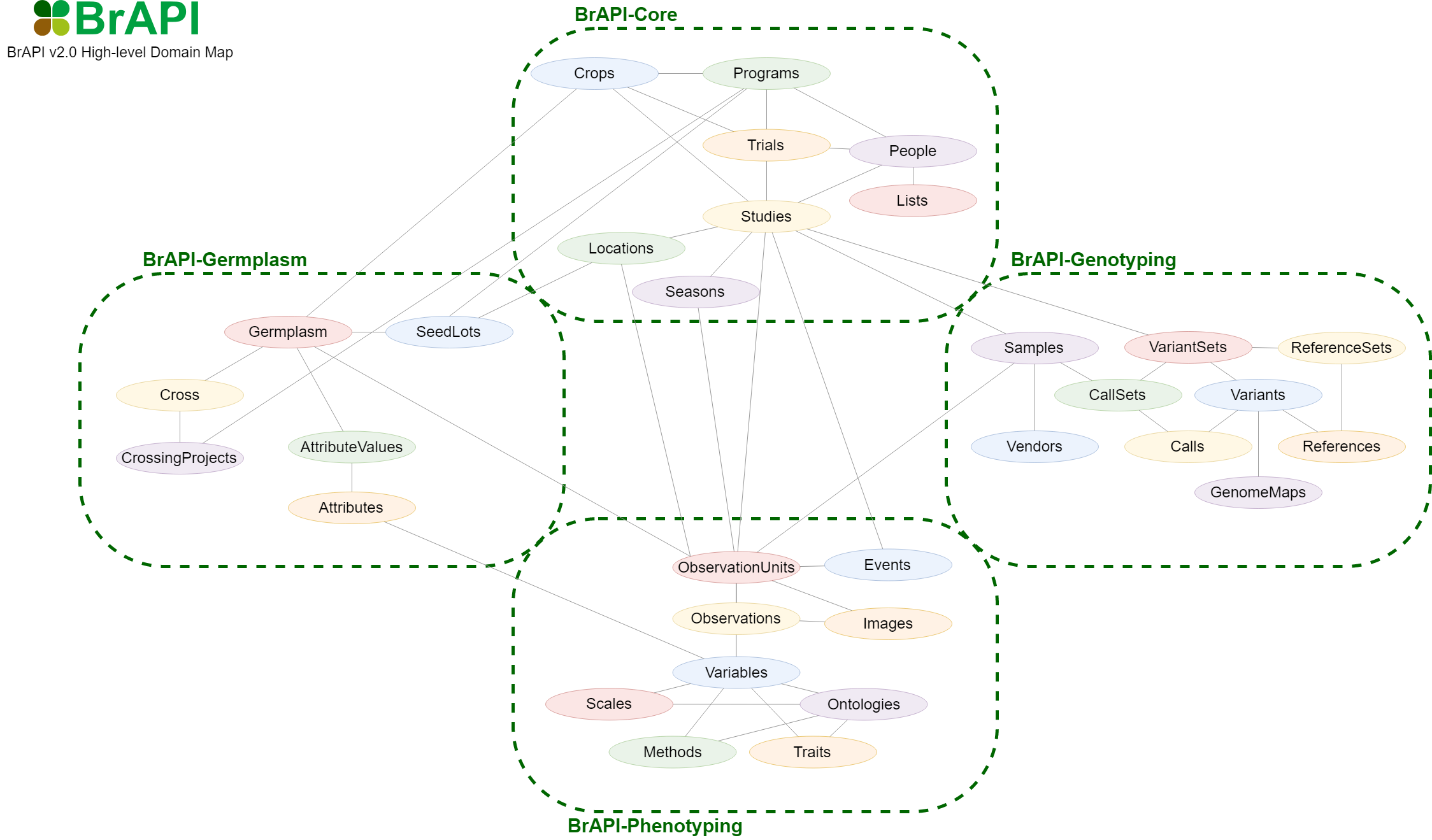
For BrAPI V1, the whole specification can be found on one page for each minor version. However, in BrAPI V2, the documentation was split across four modules: BrAPI-Core, BrAPI-Germplasm, BrAPI-Genotyping, and BrAPI-Phenotyping. These modules help organize the endpoint definitions into logical groups, but they have no impact on the implementation of BrAPI.
Project History
The BrAPI Project was initiated in June 2014, when a small group of plant breeding software teams met to discuss interoperability. This initial group consisted of representatives from Breedbase, BMS, T3, and ICRISAT. By September, they had grown to include developers from JHI, DArT, B4R, and PhenoApps. The leaders and developers from these organizations met regularly and laid the ground work for what would become the BrAPI specification. They organized the first community hackathons which have become a staple of the BrAPI community.
Over the next year, the group continued to grow with organizations like INRA, GOBii, and The Crop Ontology joining the discussion. Eventually, the group had enough web service endpoints defined that they released an initial V1 of the specification document. This initial V1 document had 36 endpoints defined focused on studies, germplasm, phenotypes, and genotype data. To this day, these categories remain as the central pillars of the BrAPI spec. In September of 2015, the GitHub repository was created and the initial spec document was copied in. This made it far easier to work simultaneously, make changes when necessary, and log issues which could be fixed later.
By 2017, it was clear to the community leaders that the project had grown so much that they needed someone working full time to manage it. About the same time, a collaborative effort between the CGIAR and the Gates Foundation formed the Excellence in Breeding (EiB) platform. One of the many goals of the EiB initiative was to hire a full time coordinator for the BrAPI project. In October of 2017, Peter Selby was hired as the first, full-time, BrAPI Project Coordinator, and he has been serving in that role since then (written as of 2022). EiB provided 5 years of funding for the project to continue to grow and establish itself, as well as facilitate data sharing between the major software systems under the CGIAR umbrella.
As the new BrAPi Project Coordinator, Peter quickly went to work building on what the community had created so far. He enforced strict code freeze and versioning guidelines, brought consistency to the existing specification, and upgraded the community website and communication streams. In 2018, versions V1.1, V1.2, and V1.3 of the specification were released, fixing many long standing technical issues and adding support for many more use cases. By the release of V1.3 in November of 2018, the specification had tripled in size to 105 active endpoints defined. These endpoints still described the same four areas of interest (studies, germplasm, phenotypes, and genotypes) but in much better detail and flexibility.